forked from p94628173/idrlnet
231 lines
8.8 KiB
Markdown
231 lines
8.8 KiB
Markdown
|
|
# Solving Simple Poisson Equation
|
||
|
|
|
||
|
|
Inspired by [Nvidia SimNet](https://developer.nvidia.com/simnet),
|
||
|
|
IDRLnet employs symbolic links to construct a computational graph automatically.
|
||
|
|
In this section, we introduce the primary usage of IDRLnet.
|
||
|
|
To solve PINN via IDRLnet, we divide the procedure into several parts:
|
||
|
|
|
||
|
|
1. Define symbols and parameters.
|
||
|
|
1. Define geometry objects.
|
||
|
|
1. Define sampling domains and corresponding constraints.
|
||
|
|
1. Define neural networks and PDEs.
|
||
|
|
1. Define solver and solve.
|
||
|
|
1. Post processing.
|
||
|
|
|
||
|
|
We provide the following example to illustrate the primary usages and features of IDRLnet.
|
||
|
|
|
||
|
|
Consider the 2d Poisson's equation defined on $\Omega=[-1,1]\times[-1,1]$, which satisfies $-\Delta u=1$, with
|
||
|
|
the boundary value conditions:
|
||
|
|
|
||
|
|
$$
|
||
|
|
\begin{align}
|
||
|
|
\frac{\partial u(x, -1)}{\partial n}&=\frac{\partial u(x, 1)}{\partial n}=0 \\
|
||
|
|
u(-1,y)&=u(1, y)=0
|
||
|
|
\end{align}
|
||
|
|
$$
|
||
|
|
|
||
|
|
## Define Symbols
|
||
|
|
For the 2d problem, we define two coordinate symbols `x` and `y`, which will be used in symbolic expressions in IDRLnet.
|
||
|
|
```python
|
||
|
|
x, y = sp.symbols('x y')
|
||
|
|
```
|
||
|
|
Note that variables `x`, `y`, `z`, `t` are reserved inside IDRLnet.
|
||
|
|
The four symbols should only represent the 4 primary coordinates.
|
||
|
|
|
||
|
|
## Define Geometric Objects
|
||
|
|
|
||
|
|
The geometry object is a simple rectangle.
|
||
|
|
```python
|
||
|
|
rec = sc.Rectangle((-1., -1.), (1., 1.))
|
||
|
|
```
|
||
|
|
|
||
|
|
Users can sample points on these geometry objects. The operators `+`, `-`, `&` are also supported.
|
||
|
|
A slightly more complicated example is as follows:
|
||
|
|
```python
|
||
|
|
import numpy as np
|
||
|
|
import idrlnet.shortcut as sc
|
||
|
|
|
||
|
|
# Define 4 polygons
|
||
|
|
I = sc.Polygon([(0, 0), (3, 0), (3, 1), (2, 1), (2, 4), (3, 4), (3, 5), (0, 5), (0, 4), (1, 4), (1, 1), (0, 1)])
|
||
|
|
D = sc.Polygon([(4, 0), (7, 0), (8, 1), (8, 4), (7, 5), (4, 5)]) - sc.Polygon(([5, 1], [7, 1], [7, 4], [5, 4]))
|
||
|
|
R = sc.Polygon([(9, 0), (10, 0), (10, 2), (11, 2), (12, 0), (13, 0), (12, 2), (13, 3), (13, 4), (12, 5), (9, 5)]) \
|
||
|
|
- sc.Rectangle(point_1=(10., 3.), point_2=(12, 4))
|
||
|
|
L = sc.Polygon([(14, 0), (17, 0), (17, 1), (15, 1), (15, 5), (14, 5)])
|
||
|
|
|
||
|
|
# Define a heart shape.
|
||
|
|
heart = sc.Heart((18, 4), radius=1)
|
||
|
|
|
||
|
|
# Union of the 5 geometry objects
|
||
|
|
geo = (I + D + R + L + heart)
|
||
|
|
|
||
|
|
# interior samples
|
||
|
|
points = geo.sample_interior(density=100, low_discrepancy=True)
|
||
|
|
plt.figure(figsize=(10, 5))
|
||
|
|
plt.scatter(x=points['x'], y=points['y'], c=points['sdf'], cmap='hot')
|
||
|
|
|
||
|
|
# boundary samples
|
||
|
|
points = geo.sample_boundary(density=400, low_discrepancy=True)
|
||
|
|
plt.scatter(x=points['x'], y=points['y'])
|
||
|
|
idx = np.random.choice(points['x'].shape[0], 400, replace=False)
|
||
|
|
|
||
|
|
# Show normal directions on boundary
|
||
|
|
plt.quiver(points['x'][idx], points['y'][idx], points['normal_x'][idx], points['normal_y'][idx])
|
||
|
|
plt.show()
|
||
|
|
```
|
||
|
|
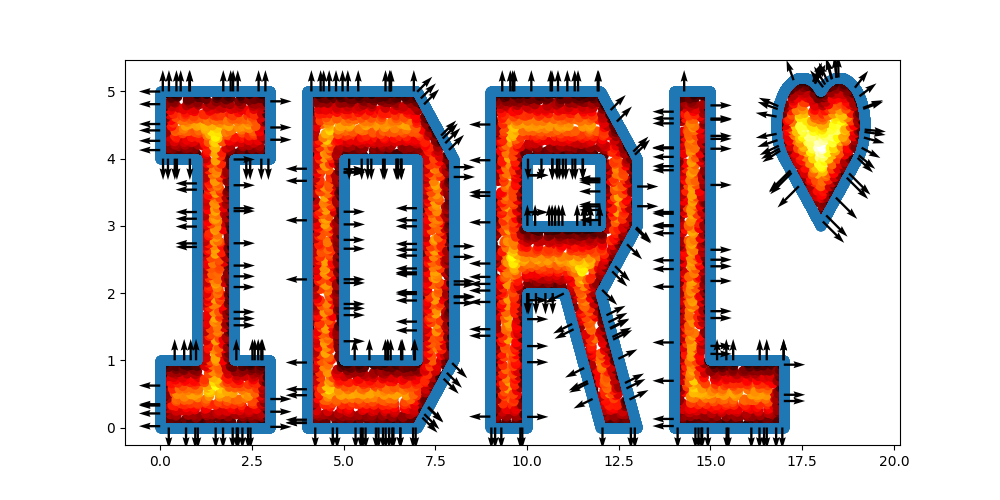
|
||
|
|
|
||
|
|
## Define Sampling Methods and Constraints
|
||
|
|
Take a 1D fitting task as an example.
|
||
|
|
The data source generates pairs $(x_i, f_i)$. We train a network $u_\theta(x_i)\approx f_i$.
|
||
|
|
Then $f_i$ is the target output of $u_\theta(x_i)$.
|
||
|
|
These targets are called constraints in IDRLnet.
|
||
|
|
|
||
|
|
For the problem, three constraints are presented.
|
||
|
|
|
||
|
|
The constraint
|
||
|
|
|
||
|
|
$$
|
||
|
|
u(-1,y)=u(1, y)=0
|
||
|
|
$$
|
||
|
|
is translated into
|
||
|
|
```python
|
||
|
|
@sc.datanode
|
||
|
|
class LeftRight(sc.SampleDomain):
|
||
|
|
# Due to `name` is not specified, LeftRight will be the name of datanode automatically
|
||
|
|
def sampling(self, *args, **kwargs):
|
||
|
|
# sieve define rules to filter points
|
||
|
|
points = rec.sample_boundary(1000, sieve=((y > -1.) & (y < 1.)))
|
||
|
|
constraints = sc.Variables({'T': 0.})
|
||
|
|
return points, constraints
|
||
|
|
```
|
||
|
|
Then `LeftRight()` is wrapped as an instance of `DataNode`.
|
||
|
|
One can store states in these instances.
|
||
|
|
Alternatively, if users do not need storing states, the code above is equivalent to
|
||
|
|
```python
|
||
|
|
@sc.datanode(name='LeftRight')
|
||
|
|
def leftright(self, *args, **kwargs):
|
||
|
|
points = rec.sample_boundary(1000, sieve=((y > -1.) & (y < 1.)))
|
||
|
|
constraints = sc.Variables({'T': 0.})
|
||
|
|
return points, constraints
|
||
|
|
```
|
||
|
|
Then `sampling()` is wrapped as an instance of `DataNode`.
|
||
|
|
|
||
|
|
The constraint
|
||
|
|
|
||
|
|
$$
|
||
|
|
\frac{\partial u(x, -1)}{\partial n}=\frac{\partial u(x, 1)}{\partial n}=0
|
||
|
|
$$
|
||
|
|
is translated into
|
||
|
|
|
||
|
|
```python
|
||
|
|
@sc.datanode(name="up_down")
|
||
|
|
class UpDownBoundaryDomain(sc.SampleDomain):
|
||
|
|
def sampling(self, *args, **kwargs):
|
||
|
|
points = rec.sample_boundary(1000, sieve=((x > -1.) & (x < 1.)))
|
||
|
|
constraints = sc.Variables({'normal_gradient_T': 0.})
|
||
|
|
return points, constraints
|
||
|
|
```
|
||
|
|
The constraint `normal_gradient_T` will also be one of the output of computable nodes, including `PdeNode` or `NetNode`.
|
||
|
|
|
||
|
|
The last constraint is the PDE itself $-\Delta u=1$:
|
||
|
|
|
||
|
|
```python
|
||
|
|
@sc.datanode(name="heat_domain")
|
||
|
|
class HeatDomain(sc.SampleDomain):
|
||
|
|
def __init__(self):
|
||
|
|
self.points = 1000
|
||
|
|
|
||
|
|
def sampling(self, *args, **kwargs):
|
||
|
|
points = rec.sample_interior(self.points)
|
||
|
|
constraints = sc.Variables({'diffusion_T': 1.})
|
||
|
|
return points, constraints
|
||
|
|
```
|
||
|
|
`diffusion_T` will also be one of the outputs of computable nodes.
|
||
|
|
`self.points` is a stored state and can be varied to control the sampling behaviors.
|
||
|
|
|
||
|
|
## Define Neural Networks and PDEs
|
||
|
|
As mentioned before, neural networks and PDE expressions are encapsulated as `Node` too.
|
||
|
|
The `Node` objects have `inputs`, `derivatives`, `outputs` properties and the `evaluate()` method.
|
||
|
|
According to their inputs, derivatives, and outputs, these nodes will be automatically connected as a computational graph.
|
||
|
|
A topological sort will be applied to the graph to decide the computation order.
|
||
|
|
|
||
|
|
```python
|
||
|
|
net = sc.get_net_node(inputs=('x', 'y',), outputs=('T',), name='net1', arch=sc.Arch.mlp)
|
||
|
|
```
|
||
|
|
This is a simple call to get a neural network with the predefined architecture.
|
||
|
|
As an alternative, one can specify the configurations via
|
||
|
|
```python
|
||
|
|
evaluate = MLP(n_seq=[2, 20, 20, 20, 20, 1)],
|
||
|
|
activation=Activation.swish,
|
||
|
|
initialization=Initializer.kaiming_uniform,
|
||
|
|
weight_norm=True)
|
||
|
|
net = NetNode(inputs=('x', 'y',), outputs=('T',), net=evaluate, name='net1', *args, **kwargs)
|
||
|
|
```
|
||
|
|
which generates a node with
|
||
|
|
- `inputs=('x','y')`,
|
||
|
|
- `derivatives=tuple()`,
|
||
|
|
- `outpus=('T')`
|
||
|
|
```python
|
||
|
|
pde = sc.DiffusionNode(T='T', D=1., Q=0., dim=2, time=False)
|
||
|
|
```
|
||
|
|
generates a node with
|
||
|
|
- `inputs=tuple()`,
|
||
|
|
- `derivatives=('T__x', 'T__y')`,
|
||
|
|
- `outputs=('diffusion_T',)`.
|
||
|
|
|
||
|
|
```python
|
||
|
|
grad = sc.NormalGradient('T', dim=2, time=False)
|
||
|
|
```
|
||
|
|
generates a node with
|
||
|
|
- `inputs=('normal_x', 'normal_y')`,
|
||
|
|
- `derivatives=('T__x', 'T__y')`,
|
||
|
|
- `outputs=('normal_gradient_T',)`.
|
||
|
|
The string `__` is reserved to represent the derivative operator.
|
||
|
|
If the required derivatives cannot be directly obtained from outputs of other nodes,
|
||
|
|
It will try `autograd` provided by Pytorch with the maximum prefix match from outputs of other nodes.
|
||
|
|
|
||
|
|
## Define A Solver
|
||
|
|
Initialize a solver to bundle all the components and solve the model.
|
||
|
|
```python
|
||
|
|
s = sc.Solver(sample_domains=(HeatDomain(), LeftRight(), UpDownBoundaryDomain()),
|
||
|
|
netnodes=[net],
|
||
|
|
pdes=[pde, grad],
|
||
|
|
max_iter=1000)
|
||
|
|
s.solve()
|
||
|
|
```
|
||
|
|
Before the solver start running, it constructs computational graphs and applies a topological sort to decide the evaluation order.
|
||
|
|
Each sample domain has its independent graph.
|
||
|
|
The procedures will be executed automatically when the solver detects potential changes in graphs.
|
||
|
|
As default, these graphs are also visualized as `png` in the `network` directory named after the corresponding domain.
|
||
|
|
|
||
|
|
The following figure shows the graph on `UpDownBoundaryDomain`:
|
||
|
|
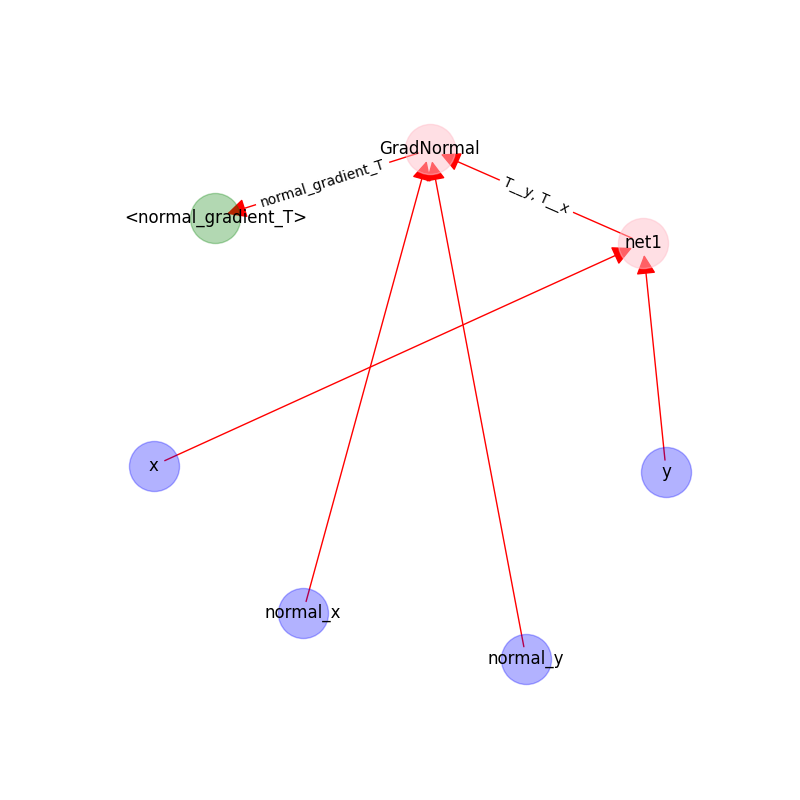
|
||
|
|
|
||
|
|
- The blue nodes are generated via sampling;
|
||
|
|
- the red nodes are computational;
|
||
|
|
- the green nodes are constraints(targets).
|
||
|
|
|
||
|
|
## Inference
|
||
|
|
We use domain `heat_domain` for inference.
|
||
|
|
First, we increase the density to 10000 via changing the attributes of the domain.
|
||
|
|
Then, `Solver.infer_step()` is called for inference.
|
||
|
|
```python
|
||
|
|
s.set_domain_parameter('heat_domain', {'points': 10000})
|
||
|
|
coord = s.infer_step({'heat_domain': ['x', 'y', 'T']})
|
||
|
|
num_x = coord['heat_domain']['x'].cpu().detach().numpy().ravel()
|
||
|
|
num_y = coord['heat_domain']['y'].cpu().detach().numpy().ravel()
|
||
|
|
num_Tp = coord['heat_domain']['T'].cpu().detach().numpy().ravel()
|
||
|
|
```
|
||
|
|
|
||
|
|
One may also define a separate domain for inference, which generates `constraints={}`, and thus, no computational graphs will be generated on the domain.
|
||
|
|
We will see this later.
|
||
|
|
|
||
|
|
## Performance Issues
|
||
|
|
1. When a domain is contained by `Solver.sample_domains`, the `sampling()` will be called every iteration.
|
||
|
|
Users should avoid including redundant domains.
|
||
|
|
Future versions will ignore domains with `constraints={}` in training steps.
|
||
|
|
2. The current version samples points in memory.
|
||
|
|
When GPU devices are enabled, data exchange between the memory and GPU devices might hinder the performance.
|
||
|
|
In future versions, we will sample points directly in GPU devices if available.
|
||
|
|
|
||
|
|
See `examples/simple_poisson`.
|